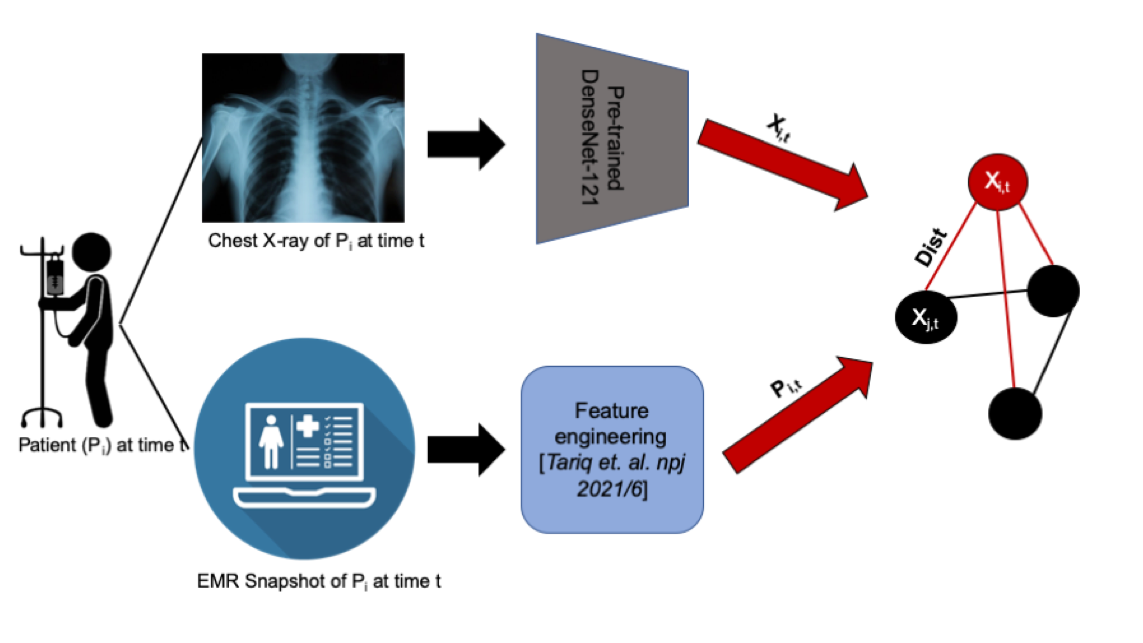
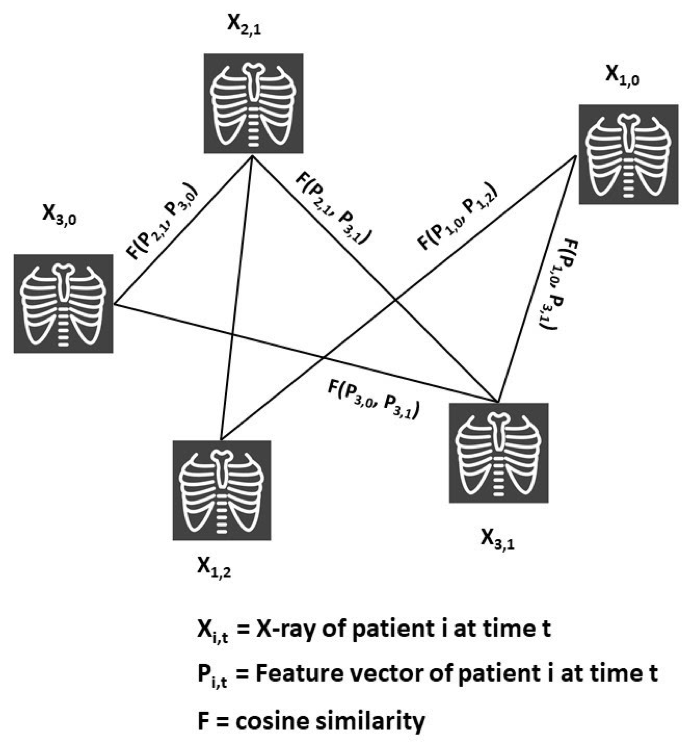
Graph-based fusion of EHR and Radiology image
To successfully perform any clinical prediction task, it is essential to learn effective representations of various data captured during patient encounters and model their interdependencies, including patient demographics, diagnostic codes, and radiologic imaging. Graph convolutional neural networks (GCN) present an intuitive and elegant way of processing multi-modal data presented as a graph structure. We proposed a novel GCN model design to understand the relationship between imaging and non-imaging data and by incorporating holistic weighted edge formation based on patient clinical history and demographic information.
- Tariq, Amara, Lin Lancaster, Praneetha Elugunti, Eric Siebeneck, Katherine Noe, Bijan Borah, James Moriarty, Imon Banerjee, and Bhavik N. Patel. “Graph convolutional network-based fusion model to predict risk of hospital acquired infections.” Journal of the American Medical Informatics Association 30, no. 6 (2023): 1056-1067.
- Tariq, Amara, Siyi Tang, Hifza Sakhi, Leo Anthony Celi, Janice M. Newsome, Daniel L. Rubin, Hari Trivedi, Judy Wawira Gichoya, and Imon Banerjee. “Fusion of imaging and non-imaging data for disease trajectory prediction for coronavirus disease 2019 patients.” Journal of Medical Imaging 10, no. 3 (2023): 034004-034004.
- Tariq, Amara, Lin Lancaster, Praneetha Elugunti, Eric Siebeneck, Katherine Noe, Bijan Borah, James Moriarty, Imon Banerjee, and Bhavik N. Patel. “Graph convolutional network-based fusion model to predict risk of hospital acquired infections.” Journal of the American Medical Informatics Association 30, no. 6 (2023): 1056-1067.
- Tang, Siyi, Amara Tariq, Jared A. Dunnmon, Umesh Sharma, Praneetha Elugunti, Daniel L. Rubin, Bhavik N. Patel, and Imon Banerjee. “Predicting 30-day all-cause hospital readmission using multimodal spatiotemporal graph neural networks.” IEEE Journal of Biomedical and Health Informatics27, no. 4 (2023): 2071-2082.
MUSE: Artificial Intelligence enabled multi-modal sensor platform for at-home health monitoring of patients with acute kidney injury (AKI)
By allowing at-home AI driven self-monitoring with easy-to-use technology, MUSE can potentially identify at-risk patients without multiple clinical visits, reduce healthcare burden and improve patient outcomes through timely assessment of risks in post-AKI patients.
Aim 1. Develop a Wearable ECG patch with analog reservoir computing
Design a battery-less patch fabricated on a soft, skin-like platform to record ECG and encode the signal using an in-sensor analog reservoir computer for ultra-low energy consumption.
Aim 2. Design a Computer-vision enabled dipstick sensor platform
Develop a single, microfluidic dipstick platform with embedded colorimetric assays for detecting four markers from urine samples through automated computer vision analysis.
Aim 3. Train a Deep learning based fusion AI model
Design a deep-learning based representation learning fusion AI model that combines multimodal sensor data (urine samples and ECG) and patient medical record (past co-morbidities and demographics) for predicting risk of adverse AKI events.
Aim 4. Smart phone application development
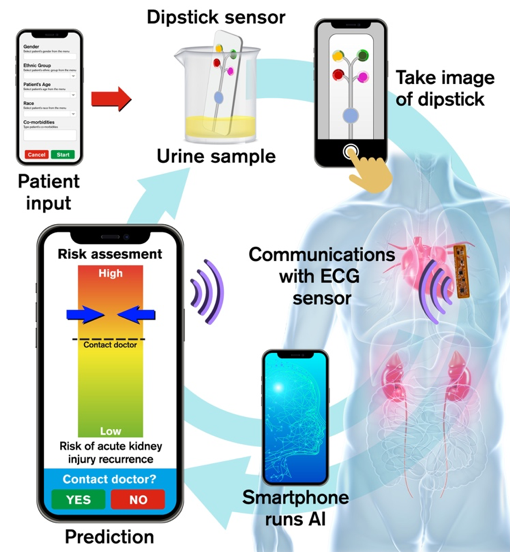
Develop a smartphone application that will: 1) allow user to input their medical record; 2) use built-in camera to capture images of dipstick platform that detects urinary bio-markers; 3) record ECG signal using near-field communication (NFC) protocol in the smartphone; 4) run computer vision and AI algorithms on-board to process sensor data, fuse with EMR for monitoring user health, and send alerts to user.
- Bhattacharya, Amartya, Sudarsan Sadasivuni, Chieh-Ju Chao, Pradyumna Agasthi, Chadi Ayoub, David R. Holmes, Reza Arsanjani, Arindam Sanyal, and Imon Banerjee. “Multi-modal fusion model for predicting adverse cardiovascular outcome post percutaneous coronary intervention.” Physiological Measurement 43, no. 12 (2022): 124004.
- Sadasivuni, Sudarsan, Sumukh Prashant Bhanushali, Imon Banerjee, and Arindam Sanyal. “In-sensor neural network for high energy efficiency analog-to-information conversion.” Scientific reports 12, no. 1 (2022): 18253.
- Sadasivuni, Sudarsan, Monjoy Saha, Neal Bhatia, Imon Banerjee, and Arindam Sanyal. “Fusion of fully integrated analog machine learning classifier with electronic medical records for real-time prediction of sepsis onset.” Scientific reports 12, no. 1 (2022): 5711.
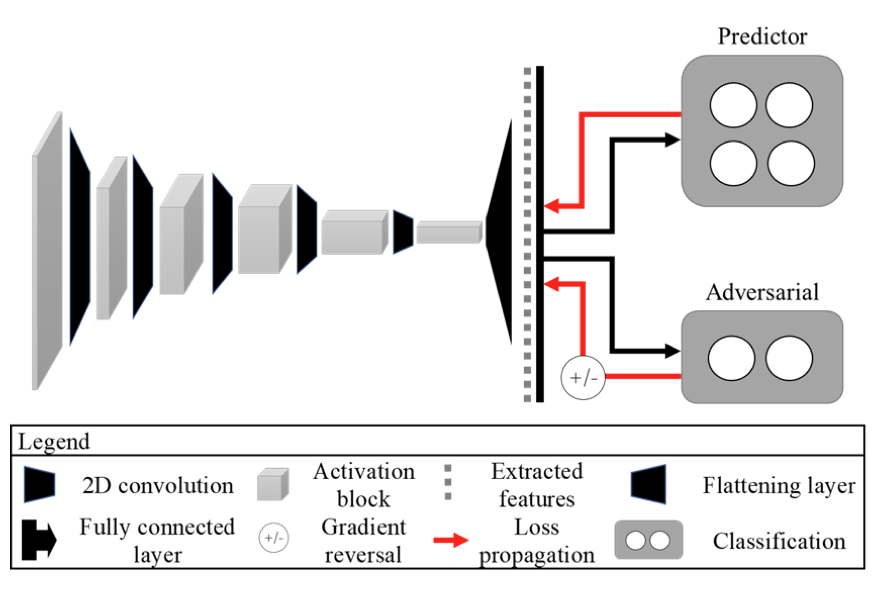
Adversarial debiasing — medical image case-study
The use of artificial intelligence (AI) in healthcare has become a very active research area in the last few years. While significant progress has been made in image classification tasks, only a few AI methods are actually being deployed in hospitals. A major hurdle in actively using clinical AI models currently is the trustworthiness of these models. More often than not, these complex models are black boxes in which promising results are generated. However, when scrutinized, these models begin to reveal implicit biases during the decision making, such as detecting race and having bias towards ethnic groups and subpopulations. In our ongoing study, we develop a two-step adversarial debiasing approach with partial learning that can reduce the racial disparity while preserving the performance of the targeted task. The methodology has been evaluated on two independent medical image case-studies – chest X-ray and mammograms, and showed promises in bias reduction while preserving the targeted performance.
- Banerjee, Imon, Kamanasish Bhattacharjee, John L. Burns, Hari Trivedi, Saptarshi Purkayastha, Laleh Seyyed-Kalantari, Bhavik N. Patel, Rakesh Shiradkar, and Judy Gichoya. ““Shortcuts” causing bias in radiology artificial intelligence: causes, evaluation and mitigation.” Journal of the American College of Radiology (2023).
- Ramasamy, Gokul, Bhavik N. Patel, and Imon Banerjee. “Anomaly Detection using Cascade Variational Autoencoder Coupled with Zero Shot Learning.” In Medical Imaging with Deep Learning, short paper track. 2023.
- Correa-Medero, Ramon L., Bhavik Patel, and Imon Banerjee. “Adversarial Debiasing techniques towards ‘fair’skin lesion classification.” In 2023 11th International IEEE/EMBS Conference on Neural Engineering (NER), pp. 1-4. IEEE, 2023.
- Correra, Ramon, Jiwoong Jason Jeong, Bhavik Patel, Hari Trivedi, Judy W. Gichoya, and Imon Banerjee. “A robust two-step adversarial debiasing with partial learning: medical image case-studies.” In Medical Imaging 2023: Imaging Informatics for Healthcare, Research, and Applications, vol. 12469, pp. 31-38. SPIE, 2023.
AI Technique for curating Large Cancer Databases (AI-LAD)
Age, racial/ethnic, and socioeconomic disparities in breast cancer treatment and survival have been widely documented for several decades and persist despite the recent advances in treatments. However, attempts to explain persistent disparities have mostly been limited to discussion of differences in insurance coverage, and on the clinical staging of the tumor and usually only include patients from a single healthcare center limited within a confined geolocation. This is primarily due to the limited availability of large-scale breast cancer patient data with long-term clinical and patient-centered outcomes to perform deep analysis of possible casual and confounding factors to study the disparities in a large scale. While multiple studies suggest that factors impacting breast cancer related morbidity and survival depend on adherence to long-term treatment (e.g. endocrine therapy), such data is only available at a smaller scale or from a single center. In order to extensively study disparities in breast cancer outcomes, the “key” is to collect a large breast cancer database of a diverse patient population by curating all the long-term clinical outcomes across longitudinal patient visits. Curating such a large database manually would not be feasible given the size and complexity of the task, and the need for multi-modal data integration, which requires hours of expert-level curation. Population-based U.S. cancer registries, such as SEER registries, are funded to collect data only on the first course of cancer therapy and cannot conduct continuous follow-up by reviewing clinical encounter notes that are necessary to capture long-term clinical outcomes, such as cancer recurrence.
Our multidisciplinary team includes computer scientists, oncologists, epidemiologists, and radiologists, and we have a strong history of collaboration in developing automated cancer informatics tools for curating long-term clinical and patient-centered outcomes data and in developing natural language processing (NLP) methods for extracting relevant information about patients from electronic medical records data. In terms of NLP, we developed AI methods that use data from free-text clinician notes, pathology and radiology reports to classify cancer recurrence status (15 best paper in cancer informatics 2019) and the sites of recurrence. We also created a weakly supervised NLP approach for extracting patient-centered outcomes for prostate cancer patients which outperformed a pre-existing rule-based model developed by the expert nursing team. We are building a flexible NLP toolset (AI-LAD) that can be executed locally at the institution level and will curate the clinical and patient-centered outcomes of breast cancer patients by parsing the clinic notes, radiology and pathology reports.
- Gupta, Anupama, Imon Banerjee, and Daniel L. Rubin. “Automatic information extraction from unstructured mammography reports using distributed semantics.” Journal of biomedical informatics 78 (2018): 78-86.
- Banerjee, Imon, Selen Bozkurt, Jennifer Lee Caswell-Jin, Allison W. Kurian, and Daniel L. Rubin. “Natural language processing approaches to detect the timeline of metastatic recurrence of breast cancer.” JCO clinical cancer informatics 3 (2019): 1-12.
- Banerjee, Imon, Kevin Li, Martin Seneviratne, Michelle Ferrari, Tina Seto, James D. Brooks, Daniel L. Rubin, and Tina Hernandez-Boussard. “Weakly supervised natural language processing for assessing patient-centered outcome following prostate cancer treatment.” JAMIA open 2, no. 1 (2019): 150-159.
Prediction of clinical event by analyzing longitudinal EHR data
Unstructured medical data analysis and integration of multimodal data (image + EHR) can unlock the large amount of electronic healthcare records (EHR) for clinical event prediction (e.g. ER visits, hospitalization, short-term mortality). Our research interest is multimodal clinical data integration and predictive modeling to closely mimic physicians workflow.
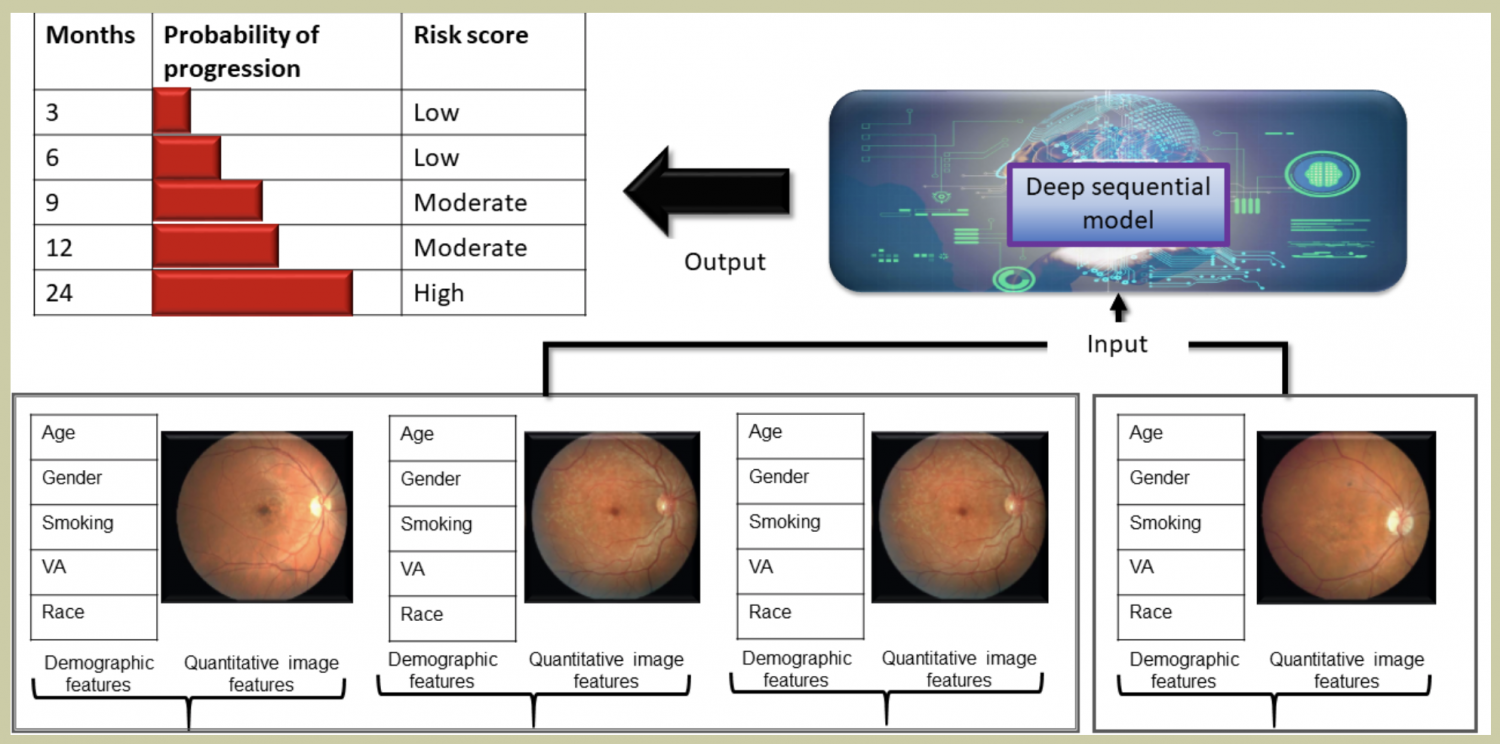
- Design a temporal deep learning model for estimating short-term life expectancy of the patients by analyzing free-text clinical notes.
- Developed a computerized technique for assessing treatment response to neoadjuvant chemotherapy by analyzing noninvasive DCE-MRI scans.
- Proposed a framework of data analysis tools for the automatic computation of qualitative and quantitative parameters to support effective annotation of patient-specific follow-up data.
- Developing a longitudinal machine learning approach to predict weight gain/loss in the context of insulin sensitivity and resistance by combining multiple omics data.
Papers/products
1. “Patient-specific COVID-19 resource utilization prediction using fusion AI model.” NPJ digital medicine 4, no. 1 (2021): 1-9. [link]
2. “Probabilistic Prognostic Estimates of Survival in Metastatic Cancer Patients (PPES-Met) Utilizing Free-Text Clinical Narratives.” [link]
3. “Assessing treatment response in triple-negative breast cancer from quantitative image analysis in perfusion magnetic resonance imaging.” [link]
4. “Integrative Personal Omics Profiles during Periods of Weight Gain and Loss.”, [link]
5. “Semantic annotation of 3D anatomical models to support diagnosis and follow-up analysis of musculoskeletal pathologies.” [link]
Margin-aware Anomaly detection for medical images
Traditional anomaly detection methods focus on detecting inter-class variations while medical image novelty identification is inherently an intra-class detection problem. For example, a machine learning model trained with normal chest X-ray and common lung abnormalities, is expected to discover and flag idiopathic pulmonary fibrosis which a rare lung disease and unseen by the model during training. The nuances from intra-class variations and lack of relevant training data in medical image analysis pose great challenges for existing anomaly detection methods. To tackle the challenges, we propose a hybrid model – nonlinear Transformation-based Embedding learn- ing for Novelty Detection (TEND). Without any out-of-distribution training data, TEND performs novelty identification by unsupervised learning of in-distribution embeddings with a vanilla AutoEncoder in the first stage and dis- criminative learning of in-distribution data and the non-linearly transformed counterparts with a binary classifier and a margin-aware objective metric in the second stage. The binary discriminator learns to distinguish the in-distribution data from the generated counterparts and outputs a class probability. The margin-aware objective is optimized jointly to include the in-distribution data in a hypersphere with a pre-defined margin and exclude the unexpected data. Even- tually, the weighted sum of class probability and the distance to margin constitutes the anomaly score.
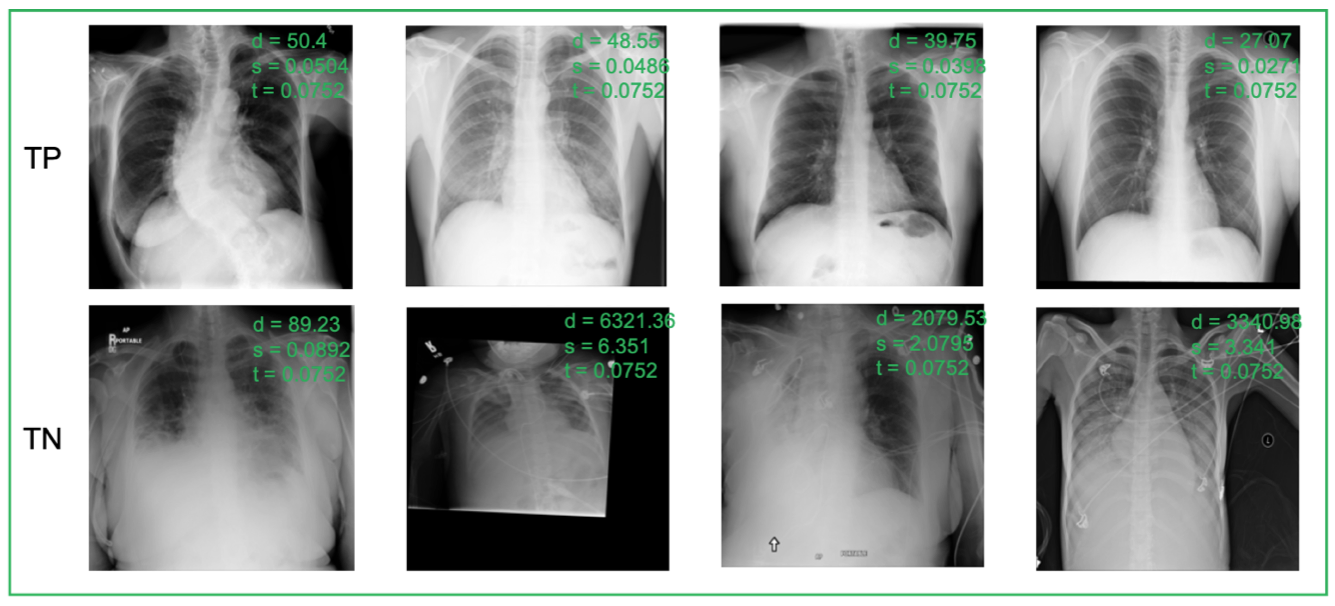
True Positive (TP, 1st row), True Negative (TN, 2nd row) predictions of TEND 500 on RSNA datasets. d: distance value from the margin learner module, p: probability outputted by the binary discriminator module, s: final score, t: optimal threshold
- Guo, Xiaoyuan, Judy Wawira Gichoya, Hari Trivedi, Saptarshi Purkayastha, and Imon Banerjee. “MedShift: Automated Identification of Shift Data for Medical Image Dataset Curation.” IEEE Journal of Biomedical and Health Informatics(2023).
- Ramasamy, Gokul, Bhavik N. Patel, and Imon Banerjee. “Anomaly Detection using Cascade Variational Autoencoder Coupled with Zero Shot Learning.” In Medical Imaging with Deep Learning, short paper track. 2023.
- Guo, Xiaoyuan, Judy Wawira Gichoya, Saptarshi Purkayastha, and Imon Banerjee. “CVAD: An Anomaly Detector for Medical Images Based on Cascade VAE.” In Workshop on Medical Image Learning with Limited and Noisy Data, pp. 187-196. Cham: Springer Nature Switzerland, 2022.
Quantitative analysis of medical images to support diagnosis.
Interested in developing computational methods that can extract quantitative information from images, integrate diverse clinical and imaging data, enable discovery of image biomarkers, and improve clinical treatment decisions. I am leading several innovative medical image analysis research projects related to cancer diagnosis, e.g. prostate cancer aggressiveness detection, histopathologic subtype classification of brain tumor, prediction of semantic features of bone tumor. I am developing a novel computational framework that can automatically interpret implicit semantic content from multimodal and/or multiparametric radiology images to enable biomedical discovery and to guide physicians in personalized care. I am responsible for the overall design of the framework, and development, execution, verification, and validation of the systems.
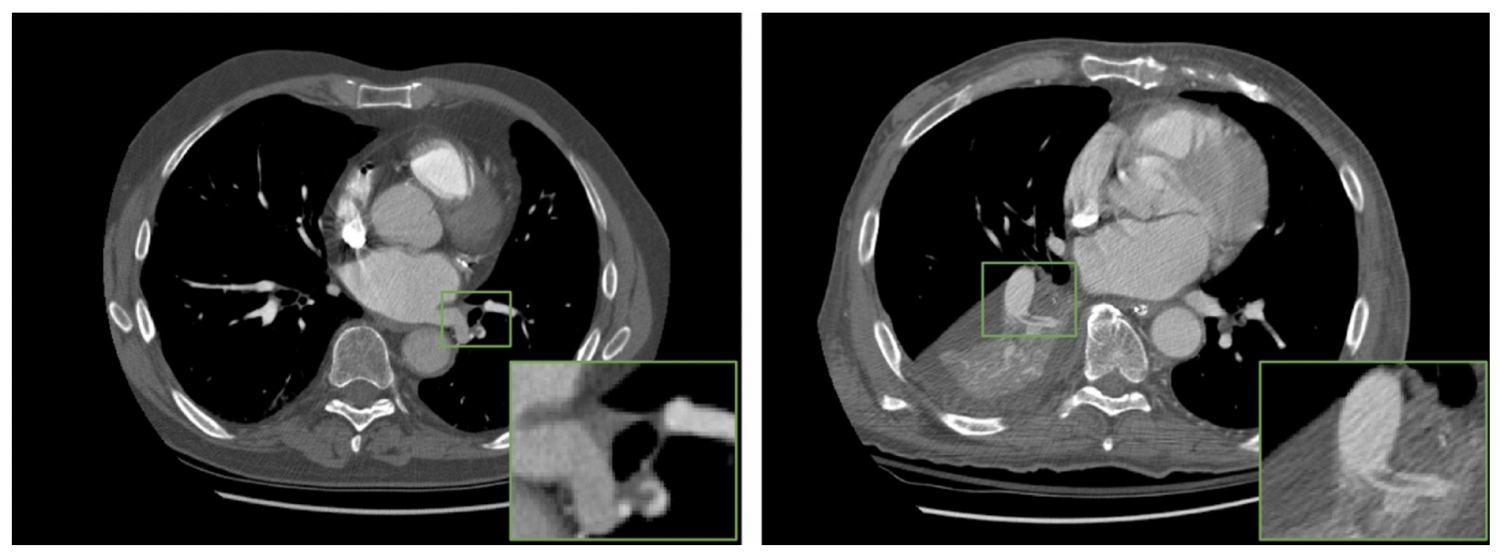
Publications and open-source code:
1. “Multimodal fusion with deep neural networks for leveraging CT imaging and electronic health record: a case-study in pulmonary embolism detection”, [link]
2. “Transfer Learning on Fused Multiparametric MR Images for Classifying Histopathological Subtypes of Rhabdomyosarcoma”, [link].
3. “Relevance feedback for enhancing content based image retrieval and automatic prediction of semantic image features: Application to bone tumor radiographs.” [link].
4. “Computerized Prediction of Radiological Observations based on Quantitative Feature Analysis: Initial Experience in Liver Lesions” [link].
5. “Computerized Multiparametric MR image Analysis for Prostate Cancer Aggressiveness-Assessment”.
Fusion of Fully Integrated Analog Machine Learning Classifier with Electronic Medical Records
The objective of this work is to develop a fusion artificial intelligence (AI) model that combines patient electronic medical record (EMR) and physiological sensor data to accurately predict early risk of sepsis and cardiovascular event. The fusion AI model has two components – an on-chip AI model that continuously analyzes patient electrocardiogram (ECG) data and a cloud AI model that combines EMR and prediction scores from on-chip AI model to predict risk score. The on-chip AI model is designed using analog circuits for sepsis prediction with high energy efficiency for integration with resource constrained wearable device. Combination of EMR and sensor physiological data improves prediction performance compared to EMR or physiological data alone, and the late fusion model has an accuracy of 93% in predicting sepsis 4 hours before onset. The key differentiation of this work over existing sepsis prediction literature is the use of single modality patient vital (ECG) and simple demographic information, instead of comprehensive laboratory test results and multiple vital signs. Such simple configuration and high accuracy makes our solution favorable for real-time, at-home use for self-monitoring.
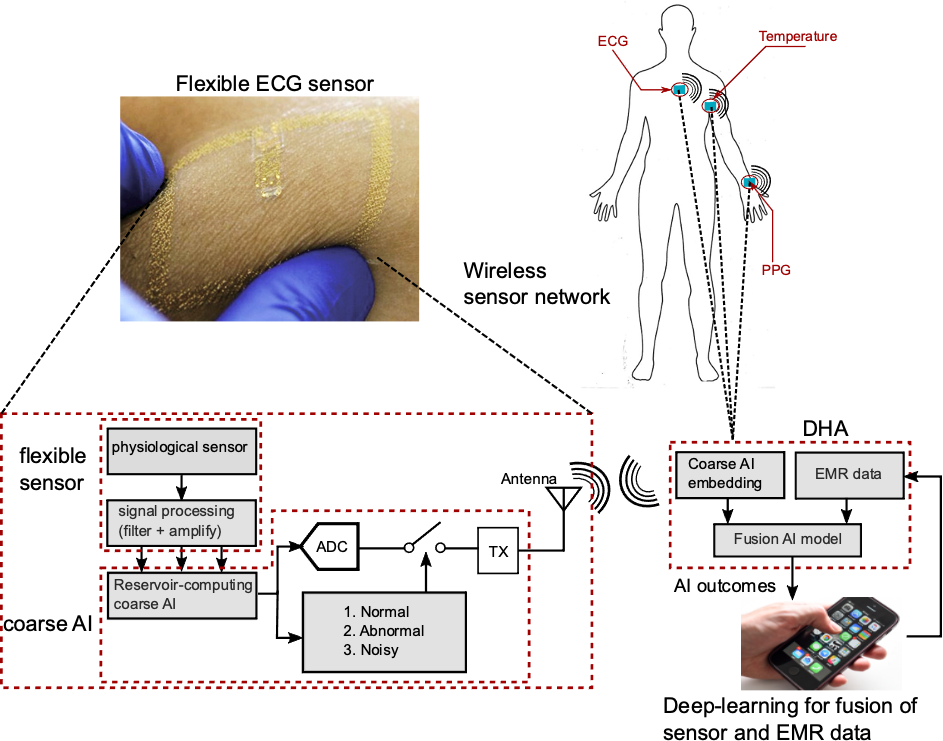
Publications and open-source code:
1. “Recurrent Neural Network Circuit for Automated Detection of Atrial Fibrillation from Raw ECG.” In 2021 IEEE International Symposium on Circuits and Systems (ISCAS), 2021. [link]
2. “Fully Integrated Analog Machine Learning Classifier Using Custom Activation Function for Low Resolution Image Classification,” [link][code]
3. “Digital Machine Learning Circuit for Real-Time Stress Detection from Wearable ECG Sensor.” [link][code]
Natural Language Processing on clinical notes
The lack of labeled data creates a data “bottleneck” for developing deep learning models for medical imaging. However, healthcare institutions have millions of imaging studies which are associated with unstructured free text radiology reports that describe imaging features and diagnoses, but there are no reliable methods for leveraging these reports to create structured labels for training deep learning models. Unstructured free text thwarts machine understanding, due to the ambiguity and variations in language among radiologists and healthcare organizations.
My research is focused in developing methods to extract structured annotations of medical images from radiology reports for training complex deep learning models.
Our method has outperformed many existing NLP algorithms on several radiology report annotation tasks (CT reports, mammography reports, US reports, and X-ray reports), as well as can infer targeted information from heterogeneous clinical notes (e.g., hospital notes, discharge summary, progress notes).
Publications and open-source code:
1. “Weakly supervised temporal model for prediction of breast cancer distant recurrence.” [link][code]
2. “Radiology Report Annotation using Intelligent Word Embeddings: Applied to Multi-institutional Chest CT Cohort,” [link][code]
3. “Weakly supervised natural language processing for assessing patient-centered outcome following prostate cancer treatment.” [link][code]
4. “Comparative effectiveness of convolutional neural network (CNN) and recurrent neural network (RNN) architectures for radiology text report classification.” [link][code available upon request]
5. A Scalable Machine Learning Approach for Inferring Probabilistic US-LI-RADS Categorization [link][code available upon request]
6. “Development and Use of Natural Language Processing for Identification of Distant Cancer Recurrence and Sites of Distant Recurrence Using Unstructured Electronic Health Record Data.” [link]

